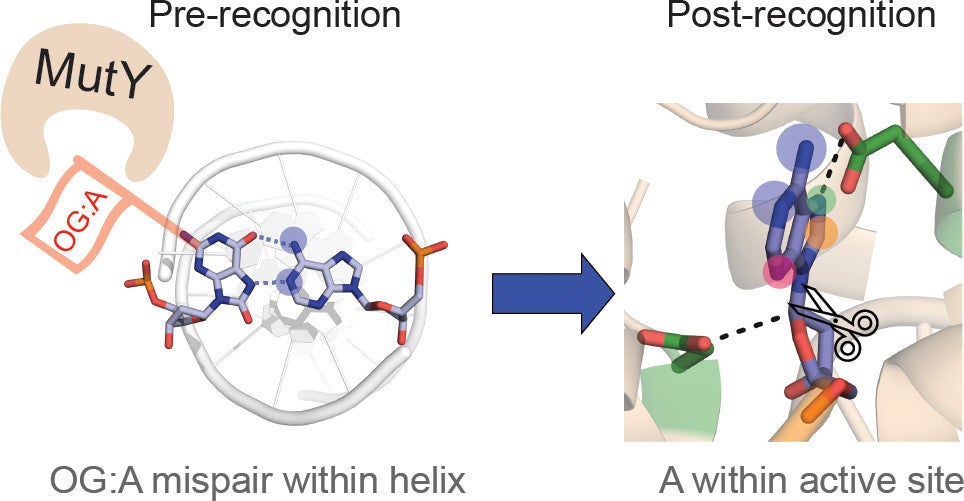
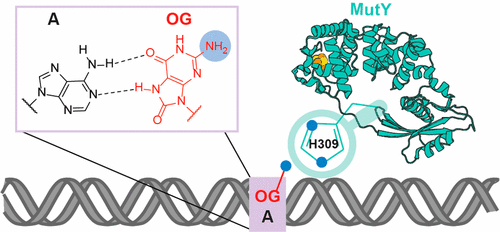
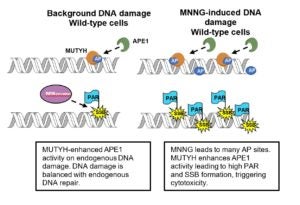
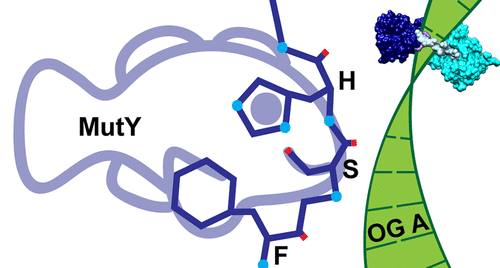
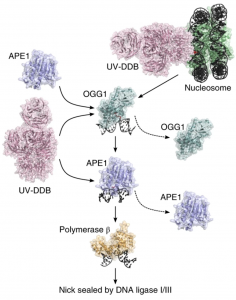
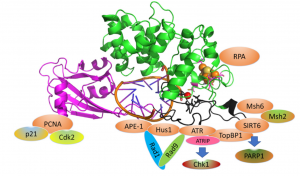
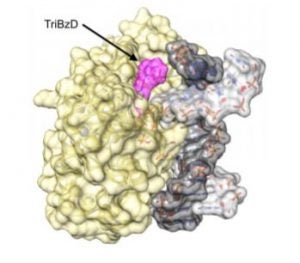
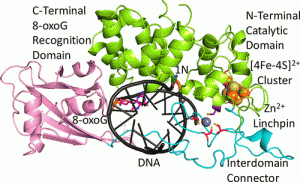
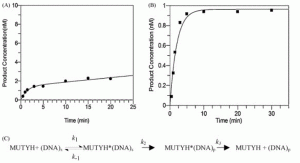
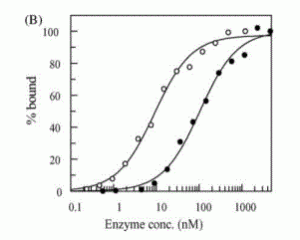
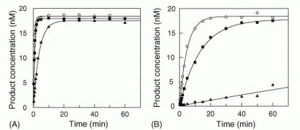
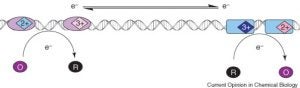
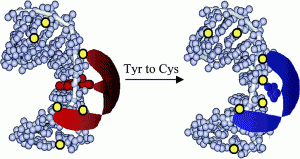
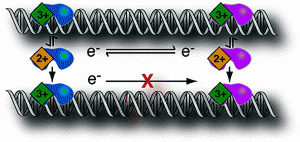
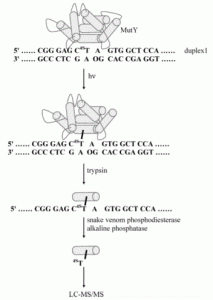
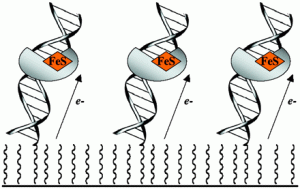
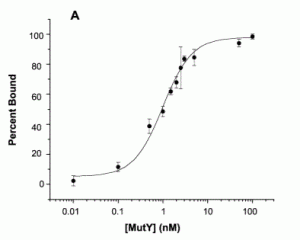
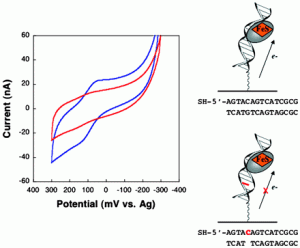
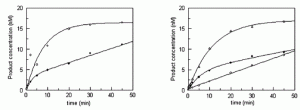
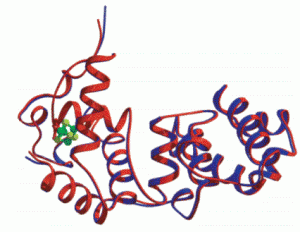
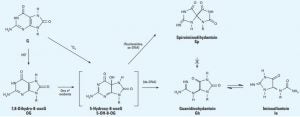
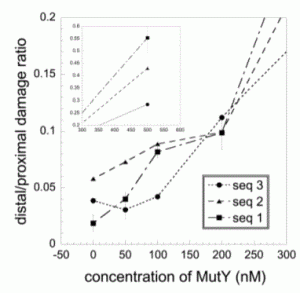
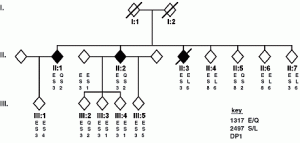
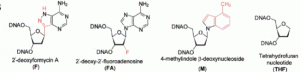
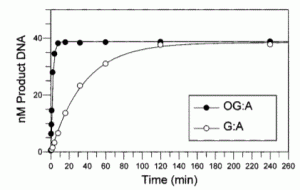
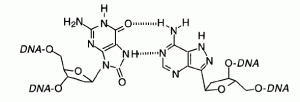
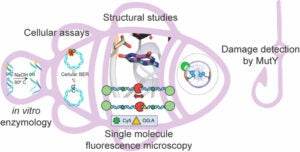
David Lab Publications:
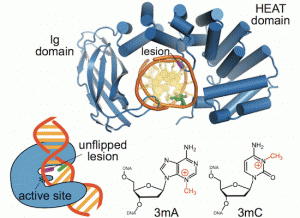
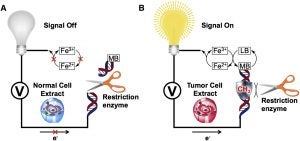
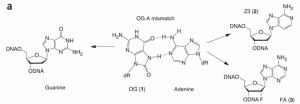
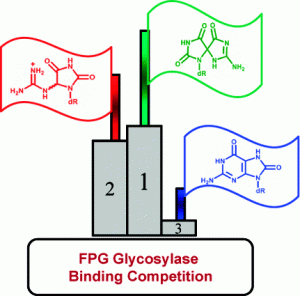
Majumdar, C.; Demir, M.; Merrill, S. R.; Hashemian, M.; David, S. S. FSHing for DNA Damage: Key Features of MutY Detection of 8-Oxoguanine:Adenine Mismatches. Acc. Chem. Res. March 2024.
https://doi.org/10.1021/acs.accounts.3c00759
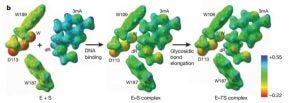
Dissanayake, U. C.; Trasviña-Arenas, C. H.; David, S. S.; Cisneros, G. A. Computational Investigation of Cancer-Associated Mutations on the Structural and Dynamical Behavior of DNA Glycosylase Enzyme (MUTY). Biophysical Journal February 2024.
https://doi.org/10.1016/j.bpj.2023.11.947
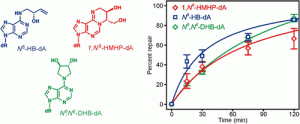
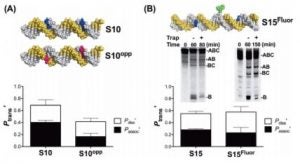
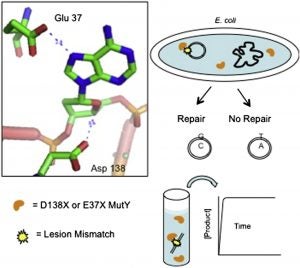
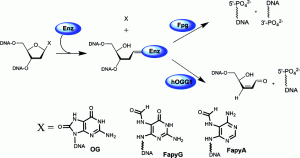
Conlon, S. G.; Khuu, C.; Trasviña-Arenas, C. H.; Xia, T.; Hamm, M. L.; Raetz, A. G.; David, S. S. Cellular Repair of Synthetic Analogs of Oxidative DNA Damage Reveals a Key Structure–Activity Relationship of the Cancer-Associated MUTYH DNA Repair Glycosylase. ACS Cent. Sci. January 2024.
https://doi.org/10.1021/acscentsci.3c00784
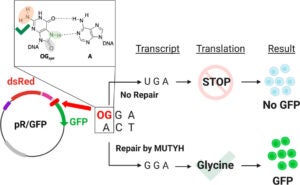
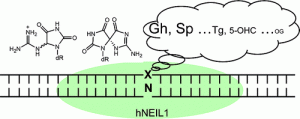
Eckenroth, B. E.; Bumgarner, J. D.; Matsumoto-Elliott, O.; David, S. S.; Doublié, S. Structural and Biochemical Insights into NEIL2’s Preference for Abasic Sites. Nucleic Acid Res. December 2023.
https://doi.org/10.1093/nar/gkad1075

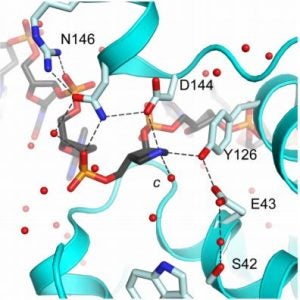
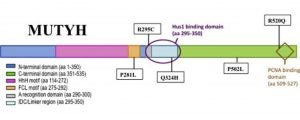
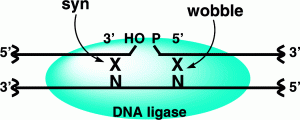
Demir, M.; Russelburg, L.P.; Lin, W.-J.; Trasviña-Arenas, C.H.; Huang, B.; Yuen, P.K.; Horvath, M.P.; David, S.S. Structural snapshots of base excision by the cancer-associated variant MutY N146S reveal a retaining mechanism. Nucleic Acid Res. January 2023.
https://doi.org/10.1093/nar/gkac1246
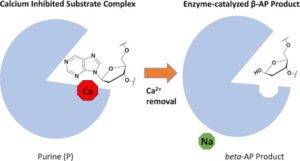
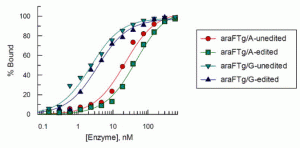
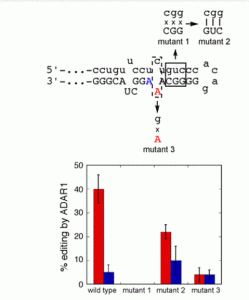
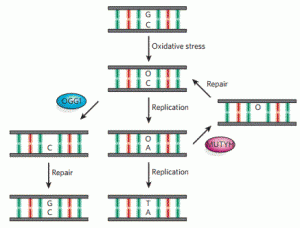
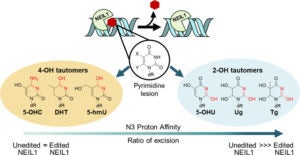
Lotsof, E.R.; Krajewski, A.E.; Anderson-Steele, B.; Rogers, J.; Zhang, L.; Yeo, J.; Conlon, S.G.; Manlove, A.H.; Lee, J.K.; David, S.S. NEIL1 Recoding due to RNA Editing Impacts Lesion-Specific Recognition and Excision. J. Am. Chem. Soc. August 2022, 144 (32), 14578-14589
https://doi.org/10.1021/jacs.2c03625
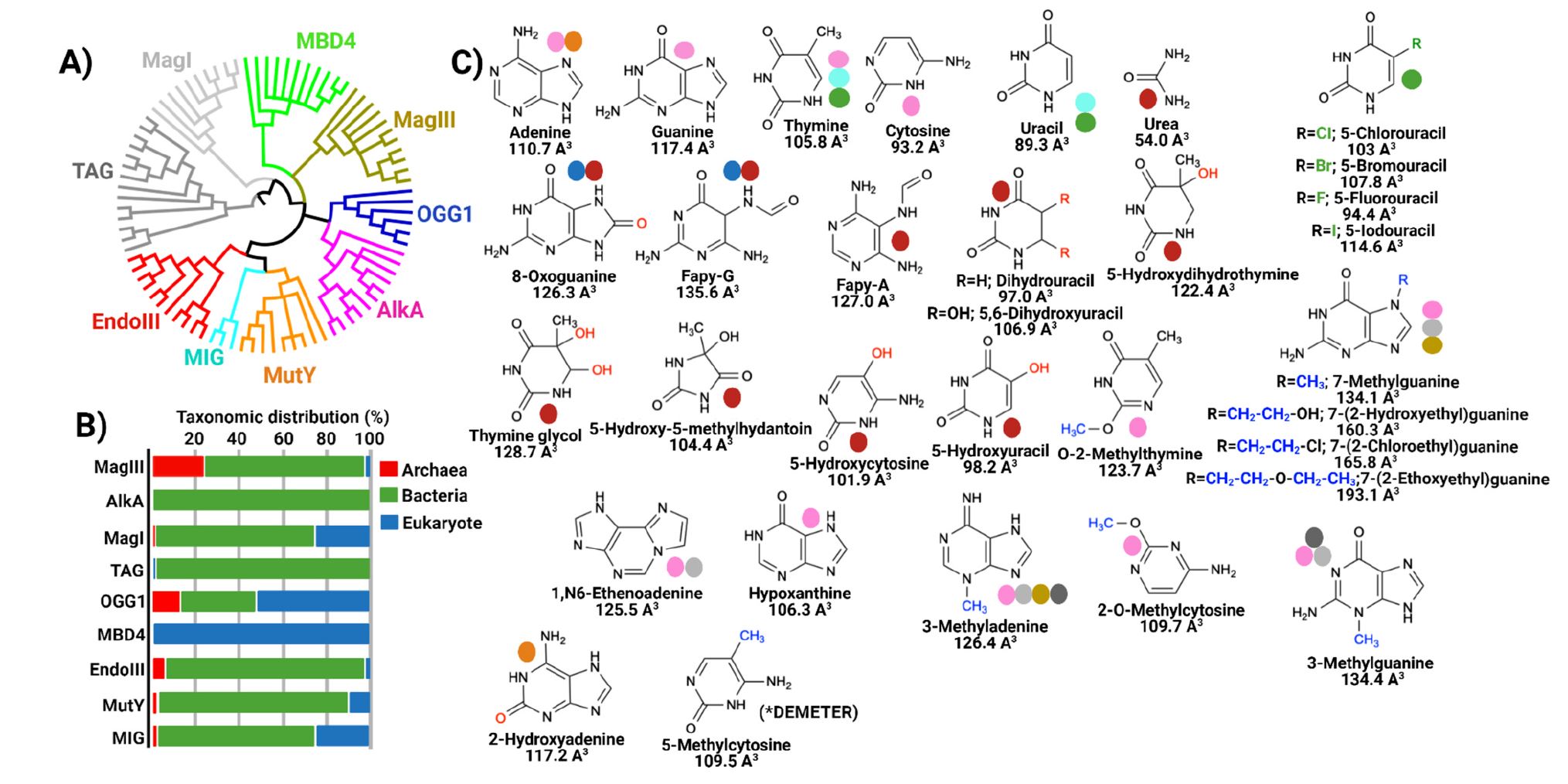
Trasviña-Arenas, C.H.; Demir, M.; Lin, W.-J.; David, S.S. Structure, function and evolution of the Helix-hairpin-Helix DNA glycosylase superfamily; Piecing together the evolutionary puzzle of DNA base damage repair mechanisms. DNA Repair. December 2021, 108, 103231.
https://doi.org/10.1016/j.dnarep.2021.103231
Majumdar, C.; Mckibbin, P.L.; Krajewski, A.E.; Manlove, A.H.; Lee, J.K.; David, S.S. Unique Hydrogen Bonding of Adenine with the Oxidatively Damaged Base 8-Oxoguanine Enables Specific Recognition and Repair by DNA Glycosylase MutY. J. Am. Soc. November 17, 2020.
https://pubs.acs.org/doi/abs/10.1021/jacs.0c06767#
Zhu, R.-Y.; Majumdar, C.; Khuu, C.; De Rosa, M.; Opresko, P. L.; David, S. S.; Kool, E. T. Designer Fluorescent Adenines Enable Real-Time Monitoring of MUTYH Activity. ACS Cent. Sci. August 31, 2020.
https://pubs.acs.org/doi/10.1021/acscentsci.0c00369
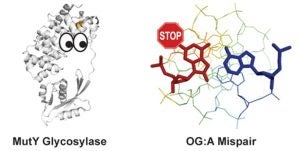
Articles ASAP, Communication: Lee, A.J.; Majumdar, C.; Kathe, S.D.; Van Ostrand, R.P.; Vickery, H.R.; Averill, A.M.; Nelson, S.R.; Manlove, A.H.; McCord, M.A.; David, S.S. Detection of OG:A Lesion Mispairs by MutY Relies on a Single His Residue and the 2-Amino Group of 8-Oxoguanine. J. Am. Chem. Soc. July 14, 2020.
https://pubs.acs.org/doi/pdf/10.1021/jacs.0c04284
Recently Published:
Raetz, A.G.; Banda, D.M.; Ma, X.; Xu, G.; Rajavel, A.N.; McKibbin, P.L.; Lebrilla, C.B.; David, S.S. The DNA repair enzyme MUTYH potentiates cytotoxicity of the alkylating agent MNNG by interacting with abasic sites. J. Biol. Chem. 2020.
Cao, S.; Rogers, JP.; Yeo, J.; Anderson-Steele, B.; Ashby, J.; David, S.S.* 2′-Fluorinated Hydantoins as Chemical Biology Tools for Base Excision Repair Glycosylases. ACS Chem. Biol. 2020. 15, 915–924.
https://pubs.acs.org/articlesonrequest/AOR-CAxYgJDa97s9meX6mcz2
Russelburg, L.P.; O’Shea Murray, V.L.; Demir, M.; Knutsen, K.R.; Sehgal, S.L.; Cao, S.; David, S.S.; Horvath, M.P. Structural basis for finding OG lesions and avoiding undamaged G by the DNA glycosylase MutY. ACS Chem. Biol. 2019. DOI: 10.1021/acschembio.9b00639.
https://pubs.acs.org/doi/pdf/10.1021/acschembio.9b00639
Jang, S.; Kumar, N.; Beckwitt, E.C.; Kong, M.; Fouquerel, E.; Rapic-Otrin, V.; Prasad, R.; Watkins, S.C.; Khuu, C.; Majumdar, C.; David, S.S.; Wilson, S.H.; Bruchez, M.P.; Opresko, P.L.; Van Houten, B. Damage sensor role of UV-DDB during base excision repair. Nat. Struct. Mol. Biol. 2019, 26, 695–703.
https://doi.org/10.1038/s41594-019-0261-7
Raetz, A.G.; David, S.S. When you’re strange: Unusual features of the MUTYH glycosylase and implications in cancer. DNA Repair. 2019, 80, 16-25.
https://doi.org/10.1016/j.dnarep.2019.05.005
Yuen, P.K.; Green, S.A.; Ashby, J.; Lay, K.T.; Santra, A.; Chen, X.; Horvath, M.P.; David, S.S. Targeting Base Excision Repair Glycosylases with DNA containing Transition State Mimics prepared via Click Chemistry. ACS Chem. Biol. 2018, DOI: 10.1021/acschembio.8b00771.
https://pubs.acs.org/doi/10.1021/acschembio.8b00771.
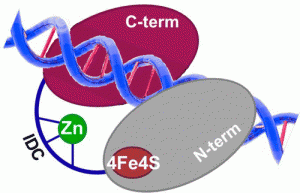
Nuñez, N.N.; Khuu, C.; Babu, C.S.; Bertolani, S.J.; Rajavel, A.N.; Spear, J.E.; Armas, J.A.; Wright, J.D.; Siegel, J.B.; Lim, C.; David, S.S. The Zinc Linchpin Motif in the DNA Repair Glycosylase MUTYH: Identifying the Zn2+ Ligands and Roles in Damage Recognition and Repair. J. Am. Chem. Soc. 2018, 140, 13260-13271.
https://pubs.acs.org/doi/abs/10.1021/jacs.8b06923.
Shi, R.; Mullins, E.A.; Shen, X.-X.; Lay, K.T.; Yuen, P.K.; David, S.S.; Rokas, A.; Eichman, B.F. Selective base excision repair of DNA damage by the non-base-flipping DNA glycosylase AlkC. EMBO J. 2017, e201797833.
http://emboj.embopress.org/content/early/2017/10/20/embj.201797833.
Manlove, A.H.; McKibbin, P.L.; Doyle, E.L.; Majumdar, C.; Hamm, M.L.; David, S.S. Structure Activity Relationships Reveal Key Features of 8-Oxoguanine:A Mismatch Detection by the MutY Glycosylase. ACS Chem. Biol. 2017. 12, 2335–2344.
http://pubs.acs.org/doi/abs/10.1021/acschembio.7b00389.
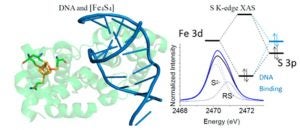
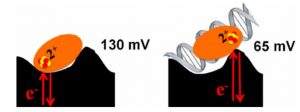
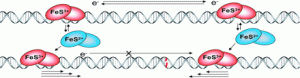
Ha, Y.; Arnold, A.R.; Nuñez, N.N.; Bartels, P.L.; Zhou, A.; David. S.S.; Barton, J.K.; Hedman, B.; Hodgson, K.O.; Solomon, E.I. S K-edge XAS Studies of the Effect of DNA Binding on the [Fe4S4] Site in EndoIII and MutY. J. Am. Chem. Soc. 2017, 139, 11434–1144.
http://pubs.acs.org/doi/abs/10.1021/jacs.7b03966.
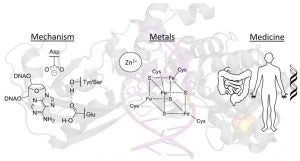
Banda, D. M.; Nuñez, N. N.; Burnside, M. A.; Bradshaw, K. M.; David, S. S., Repair of 8-oxoG:A Mismatches by the MUTYH Glycosylase: Mechanisms, Metals and Medicine. Free Radical Biol. Med. 2017, 107, 202-215. http://www.sciencedirect.com/science/article/pii/S0891584917300060.
Bartels, P. L.; Zhou, A.; Arnold, A. R.; Barton, J. K.; Nuñez, N. N.; David, S. S.; Crespilho, F. N., Electrochemistry of the [4Fe4S] Cluster in Base Excision Repair Proteins: Tuning the Redox Potential with DNA. Langmuir. 2017, 33 (10), 2523-2530. http://dx.doi.org/10.1021/acs.langmuir.6b04581.
Woods, R. D.; Chu, A.; Cao, S.; Richards, J. L.; David, S. S.; O’Shea, V. L.; Horvath, M. P., Structure and stereochemistry of the base excision repair glycosylase MutY reveal a mechanism similar to retaining glycosidases. Nucleic Acids Res. 2016, 44 (2), 801-10. http://doi.org/10.1093/nar/gkv1469.
Wickramaratne, S.; Banda, D. M.; Ji, S.; Manlove, A. H.; Malayappan, B.; Nuñez, N. N.; Samson, L.; Campbell, C.; David, S. S.; Tretyakova, N., Base Excision Repair of N6-Deoxyadenosine Adducts of 1,3-Butadiene. Biochemistry. 2016, 55 (43), 6070-6081. http://pubs.acs.org/doi/abs/10.1021/acs.biochem.6b00553.
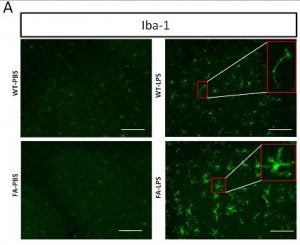
Shen, Y.; McMackin, M. Z.; Shan, Y.; Cortopassi, G.; Raetz, A.; David, S., Frataxin Deficiency Promotes Excess Microglial DNA Damage and Inflammation that Is Rescued by PJ34. PLoS One. 2016, 11 (3), e0151026. http://doi.org/10.1371/journal.pone.0151026.
Nuñez, N. N.; Manlove, A. H.; David, S. S., DNMT1 and Cancer: An Electrifying Link. Chem Biol. 2015, 22 (7), 810-1. http://doi.org/10.1016/j.chembiol.2015.07.004.
Mullins, E. A.; Shi, R.; Parsons, Z. D.; Eichman, B. F.; Yuen, P. K.; David, S. S.; Igarashi, Y., The DNA glycosylase AlkD uses a non-base-flipping mechanism to excise bulky lesions. Nature. 2015, 527 (7577), 254-8. http://doi.org/10.1038/nature15728.
Brinkmeyer, M. K.; David, S. S., Distinct functional consequences of MUTYH variants associated with colorectal cancer: Damaged DNA affinity, glycosylase activity and interaction with PCNA and Hus1. DNA Repair. 2015, 34, 39-51. https://doi.org/10.1016/j.dnarep.2015.08.001.
Rowland, M. M.; Schonhoft, J. D.; McKibbin, P. L.; David, S. S.; Stivers, J. T., Microscopic mechanism of DNA damage searching by hOGG1. Nucleic Acids Res. 2014, 42 (14), 9295-9303. https://doi.org/10.1093/nar/gku621.
Engstrom, L. M.; Brinkmeyer, M. K.; Ha, Y.; Raetz, A. G.; Hedman, B.; Hodgson, K. O.; Solomon, E. I.; David, S. S., A Zinc Linchpin Motif in the MUTYH Glycosylase Interdomain Connector Is Required for Efficient Repair of DNA Damage. J. Am. Chem. Soc. 2014, 136 (22), 7829-7832. http://dx.doi.org/10.1021/ja502942d.
McKibbin, P. L.; Fleming, A. M.; Towheed, M. A.; Van Houten, B.; Burrows, C. J.; David, S. S., Repair of Hydantoin Lesions and Their Amine Adducts in DNA by Base and Nucleotide Excision Repair. J. Am. Chem. Soc. 2013, 135 (37), 13851-13861. https://dx.doi.org/10.1021/ja4059469.
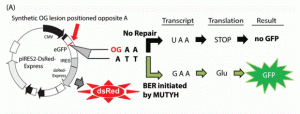
Raetz, A. G.; Xie, Y.; Kundu, S.; Brinkmeyer, M. K.; Chang, C.; David, S. S., Cancer-associated variants and a common polymorphism of MUTYH exhibit reduced repair of oxidative DNA damage using a GFP-based assay in mammalian cells. Carcinogenesis. 2012, 33 (11), 2301-2309. http://doi.org/10.1093/carcin/bgs270.
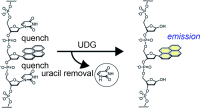
Ono, T.; Wang, S.; Koo, C.-K.; Engstrom, L.; David, S. S.; Kool, E. T., Direct fluorescence monitoring of DNA base excision repair. Angew. Chem., Int. Ed. 2012, 51 (7), 1689-1692, S1689/1-S1689/10. http://doi.org/10.1002/anie.201108135.
Onizuka, K.; Yeo, J.; David, S. S.; Beal, P. A., NEIL1 Binding to DNA Containing 2′-Fluorothymidine Glycol Stereoisomers and the Effect of Editing. ChemBioChem. 2012, 13 (9), 1338 1348. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3454477/.
Michelson, A. Z.; Rozenberg, A.; Tian, Y.; Sun, X.; Davis, J.; Francis, A. W.; O’Shea, V. L.; Halasyam, M.; Manlove, A. H.; David, S. S.; Lee, J. K., Gas-Phase Studies of Substrates for the DNA Mismatch Repair Enzyme MutY. J. Am. Chem. Soc. 2012, 134 (48), 19839-19850. https://dx.doi.org/10.1021/ja309082k.
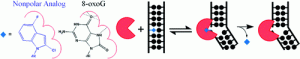
McKibbin, P. L.; Kobori, A.; Taniguchi, Y.; Kool, E. T.; David, S. S., Surprising Repair Activities of Nonpolar Analogs of 8-oxoG Expose Features of Recognition and Catalysis by Base Excision Repair Glycosylases. J. Am. Chem. Soc. 2012, 134 (3), 1653-1661. https://dx.doi.org/10.1021/ja208510m.
Engstrom, L. M.; Partington, O. A.; David, S. S., An Iron-Sulfur Cluster Loop Motif in the Archaeoglobus fulgidus Uracil-DNA Glycosylase Mediates Efficient Uracil Recognition and Removal. Biochemistry. 2012, 51 (25), 5187-5197. https://dx.doi.org/10.1021/bi3000462.
Brinkmeyer, M. K.; Pope, M. A.; David, S. S., Catalytic Contributions of Key Residues in the Adenine Glycosylase MutY Revealed by pH-dependent Kinetics and Cellular Repair Assays. Chem. Biol. (Oxford, U. K.). 2012, 19 (2), 276-286. http://doi.org/10.1016/j.chembiol.2011.11.011.
Chu, A. M.; Fettinger, J. C.; David, S. S., Profiling base excision repair glycosylases with synthesized transition state analogs. Bioorg. Med. Chem. Lett. 2011, 21 (17), 4969-4972. http://doi.org/10.1016/j.bmcl.2011.05.085.
Zhao, X.; Krishnamurthy, N.; Burrows, C. J.; David, S. S., Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry. 2010, 49 (8), 1658-1666. https://dx.doi.org/10.1021/bi901852q.
Yeo, J.; Goodman, R. A.; Schirle, N. T.; David, S. S.; Beal, P. A., RNA editing changes the lesion specificity for the DNA repair enzyme NEIL1. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (48), 20715-20719, S20715/1-S20715/4. http://doi.org/10.1073/pnas.1009231107.
Kundu, S.; Brinkmeyer, M. K.; Eigenheer, R. A.; David, S. S., Ser 524 is a phosphorylation site in MUTYH and Ser 524 mutations alter 8-oxoguanine (OG):A mismatch recognition. DNA Repair. 2010, 9 (10), 1026-1037. http://doi.org/10.1016/j.dnarep.2010.07.002.
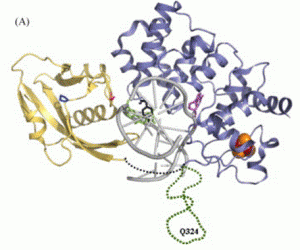
Kundu, S.; Brinkmeyer, M. K.; Livingston, A. L.; David, S. S., Adenine removal activity and bacterial complementation with the human MutY homologue (MUTYH) and Y165C, G382D, P391L and Q324R variants associated with colorectal cancer. DNA Repair. 2009, 8 (12), 1400-1410. http://doi.org/10.1016/j.dnarep.2009.09.009.
Livingston, A. L.; O’Shea, V. L.; Kim, T.; Kool, E. T.; David, S. S., Unnatural substrates reveal the importance of 8-oxoguanine for in vivo mismatch repair by MutY. Nat. Chem. Biol. 2008, 4 (1), 51-58. http://doi.org/10.1038/nchembio.2007.40.
Krishnamurthy, N.; Zhao, X.; Burrows, C. J.; David, S. S., Superior Removal of Hydantoin Lesions Relative to Other Oxidized Bases by the Human DNA Glycosylase hNEIL1. Biochemistry. 2008, 47 (27), 7137-7146. https://dx.doi.org/10.1021/bi800160s.
Krishnamurthy, N.; Haraguchi, K.; Greenberg, M. M.; David, S. S., Efficient removal of formamidopyrimidines by 8-oxoguanine glycosylases. Biochemistry. 2008, 47 (3), 1043-1050. http://pubs.acs.org/doi/abs/10.1021/bi701619u.
David, S. S.; Meggers, E., Inorganic chemical biology: from small metal complexes in biological systems to metalloproteins. Curr. Opin. Chem. Biol. 2008, 12 (2), 194-196. https://dx.doi.org/10.1016/j.cbpa.2008.03.008.
Zhao, X.; Muller, J. G.; Halasyam, M.; David, S. S.; Burrows, C. J., In vitro ligation of oligodeoxynucleotides containing C8-oxidized purine lesions using bacteriophage T4 DNA ligase. Biochemistry. 2007, 46 (12), 3734-3744. https://dx.doi.org/10.1021/bi062214k.
Krishnamurthy, N.; Muller, J. G.; Burrows, C. J.; David, S. S., Unusual Structural Features of Hydantoin Lesions Translate into Efficient Recognition by Escherichia coli Fpg. Biochemistry. 2007, 46 (33), 9355-9365. https://dx.doi.org/10.1021/bi602459v.
David, S. S.; O’Shea, V. L.; Kundu, S., Base-excision repair of oxidative DNA damage. Nature (London, U. K.). 2007, 447 (7147), 941-950. http://doi.org/10.1038/nature05978.
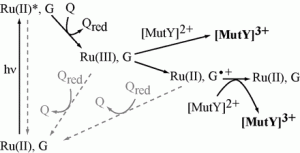
Yavin, E.; Stemp, E. D. A.; O’Shea, V. L.; David, S. S.; Barton, J. K., Electron trap for DNA-bound repair enzymes: a strategy for DNA-mediated signaling. Proc. Natl. Acad. Sci. U. S. A. 2006, 103 (10), 3610-3614. http://doi.org/10.1073/pnas.0600239103.
Yavin, E.; Boal, A. K.; Stemp, E. D. A.; Boon, E. M.; Livingston, A. L.; O’Shea, V. L.; David, S. S.; Barton, J. K., Protein-DNA charge transport: redox activation of a DNA repair protein by guanine radical. Proc. Natl. Acad. Sci. U. S. A. 2005, 102 (10), 3546-3551. http://doi.org/10.1073/pnas.0409410102.
Pope, M. A.; David, S. S., DNA damage recognition and repair by the murine MutY homologue. DNA Repair. 2005, 4 (1), 91-102. http://doi.org/10.1016/j.dnarep.2004.08.004.
Pope, M. A.; Chmiel, N. H.; David, S. S., Insight into the functional consequences of hMYH variants associated with colorectal cancer: distinct differences in the adenine glycosylase activity and the response to AP endonucleases of Y150C and G365D murine MYH. DNA Repair. 2005, 4 (3), 315-325. http://doi.org/10.1016/j.dnarep.2004.10.003.
Lukianova, O. A.; David, S. S., A role for iron-sulfur clusters in DNA repair. Curr. Opin. Chem. Biol. 2005, 9 (2), 145-151. http://doi.org/10.1016/j.cbpa.2005.02.006.
Livingston, A. L.; Kundu, S.; Henderson, P. M.; Anderson, D. W.; David, S. S., Insight into the roles of tyrosine 82 and glycine 253 in the Escherichia coli adenine glycosylase MutY. Biochemistry. 2005, 44 (43), 14179-90. http://doi.org/10.1021/bi050976u.
David, S. S., Structural biology: DNA search and rescue. Nature (London, U. K.). 2005, 434 (7033), 569-570. http://doi.org/10.1038/434569a.
Boal, A. K.; Yavin, E.; Lukianova, O. A.; O’Shea, V. L.; David, S. S.; Barton, J. K., DNA-Bound Redox Activity of DNA Repair Glycosylases Containing [4Fe-4S] Clusters. Biochemistry. 2005, 44 (23), 8397-8407. http://doi.org/10.1021/bi047494n.
Chepanoske, C. L.; Lukianova, O. A.; Lombard, M.; Golinelli-Cohen, M.-P.; David, S. S., A Residue in MutY Important for Catalysis Identified by Photocross-Linking and Mass Spectrometry. Biochemistry. 2004, 43 (3), 651-662. http://doi.org/10.1021/bi035537e.
Boon, E. M.; Livingston, A. L.; Chmiel, N. H.; David, S. S.; Barton, J. K., DNA-mediated charge transport for DNA repair. [Erratum to document cited in CA140:055464]. Proc. Natl. Acad. Sci. U. S. A. 2004, 101 (13), 4718. http://doi.org/10.1073/pnas.2035257100.
Wiederholt, C. J.; Delaney, M. O.; Pope, M. A.; David, S. S.; Greenberg, M. M., Repair of DNA Containing Fapy·dG and Its β-C-Nucleoside Analogue by Formamidopyrimidine DNA Glycosylase and MutY. Biochemistry. 2003, 42 (32), 9755-9760. http://doi.org/10.1021/bi034844h.
Leipold, M. D.; Workman, H.; Muller, J. G.; Burrows, C. J.; David, S. S., Recognition and Removal of Oxidized Guanines in Duplex DNA by the Base Excision Repair Enzymes hOGG1, yOGG1, and yOGG2. Biochemistry. 2003, 42 (38), 11373-11381. http://doi.org/10.1021/bi034951b.
Francis, A. W.; Helquist, S. A.; Kool, E. T.; David, S. S., Probing the Requirements for Recognition and Catalysis in Fpg and MutY with Nonpolar Adenine Isosteres. J. Am. Chem. Soc. 2003, 125 (52), 16235-16242. http://pubs.acs.org/doi/abs/10.1021/ja0374426.
Francis, A. W.; David, S. S., Escherichia coli MutY and Fpg Utilize a Processive Mechanism for Target Location. Biochemistry. 2003, 42 (3), 801-810. http://pubs.acs.org/doi/abs/10.1021/bi026375%2B.
Chmiel, N. H.; Livingston, A. L.; David, S. S., Insight into the Functional Consequences of Inherited Variants of the hMYH Adenine Glycosylase Associated with Colorectal Cancer: Complementation Assays with hMYH Variants and Pre-steady-state Kinetics of the Corresponding Mutated E. coli Enzymes. J. Mol. Biol. 2003, 327 (2), 431-443. http://www.sciencedirect.com/science/article/pii/S0022283603001244.
Boon, E. M.; Livingston, A. L.; Chmiel, N. H.; David, S. S.; Barton, J. K., DNA-mediated charge transport for DNA repair. Proc. Natl. Acad. Sci. U. S. A. 2003, 100 (22), 12543-12547. http://www.pnas.org/content/100/22/12543.full.
Pope, M. A.; Porello, S. L.; David, S. S., Escherichia coli apurinic-apyrimidinic endonucleases enhance the turnover of the adenine glycosylase MutY with G:A substrates. J. Biol. Chem. 2002, 277 (25), 22605-22615. http://www.jbc.org/content/277/25/22605.long.
Messick, T. E.; Chmiel, N. H.; Golinelli, M.-P.; Langer, M. R.; Joshua-Tor, L.; David, S. S., Noncysteinyl Coordination to the [4Fe-4S]2+ Cluster of the DNA Repair Adenine Glycosylase MutY Introduced via Site-Directed Mutagenesis. Structural Characterization of an Unusual Histidinyl-Coordinated Cluster. Biochemistry. 2002, 41 (12), 3931-3942. http://pubs.acs.org/doi/abs/10.1021/bi012035x.
Burrows, C. J.; Muller, J. G.; Kornyushyna, O.; Luo, W.; Duarte, V.; Leipold, M. D.; David, S. S., Structure and potential mutagenicity of new hydantoin products from guanosine and 8-oxo-7,8-dihydroguanine oxidation by transition metals. Environ. Health Perspect. Suppl. 2002, 110 (5), 713-717. https://www.ncbi.nlm.nih.gov/pubmed/12426118.
Boon, E. M.; Pope, M. A.; Williams, S. D.; David, S. S.; Barton, J. K., DNA-Mediated Charge Transport as a Probe of MutY/DNA Interaction. Biochemistry. 2002, 41 (26), 8464-8470. http://pubs.acs.org/doi/abs/10.1021/bi012068c.
Al-Tassan, N.; Chmiel, N. H.; Maynard, J.; Fleming, N.; Livingston, A. L.; Williams, G. T.; Hodges, A. K.; Davies, D. R.; David, S. S.; Sampson, J. R.; Cheadle, J. P., Inherited variants of MYH associated with somatic G:C→T:A mutations in colorectal tumors. Nat. Genet. 2002, 30 (2), 227-232. http://www.nature.com/ng/journal/v30/n2/full/ng828.html.
Chmiel, N. H.; Golinelli, M. P.; Francis, A. W.; David, S. S., Efficient recognition of substrates and substrate analogs by the adenine glycosylase MutY requires the C-terminal domain. Nucleic Acids Res. 2001, 29 (2), 553-64. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC29658/.
Williams, S. D.; David, S. S., A Single Engineered Point Mutation in the Adenine Glycosylase MutY Confers Bifunctional Glycosylase/AP Lyase Activity. Biochemistry. 2000, 39 (33), 10098-10109. http://pubs.acs.org/doi/abs/10.1021/bi0004652.
Leipold, M. D.; Muller, J. G.; Burrows, C. J.; David, S. S., Removal of Hydantoin Products of 8-Oxoguanine Oxidation by the Escherichia coli DNA Repair Enzyme, FPG. Biochemistry. 2000, 39 (48), 14984-14992. http://pubs.acs.org/doi/abs/10.1021/bi0017982.
Chepanoske, C. L.; Langelier, C. R.; Chmiel, N. H.; David, S. S., Recognition of the Nonpolar Base 4-Methylindole in DNA by the DNA Repair Adenine Glycosylase MutY. Org. Lett. 2000, 2 (9), 1341-1344. http://pubs.acs.org/doi/abs/10.1021/ol005831o.
Chepanoske, C. L.; Golinelli, M. P.; Williams, S. D.; David, S. S., Positively charged residues within the iron-sulfur cluster loop of E. coli MutY participate in damage recognition and removal. Arch Biochem Biophys. 2000, 380 (1), 11-9. http://www.sciencedirect.com/science/article/pii/S0003986100918903.
Williams, S. D.; David, S. S., Formation of a Schiff Base Intermediate Is Not Required for the Adenine Glycosylase Activity of Escherichia coli MutY. Biochemistry. 1999, 38 (47), 15417-15424. http://pubs.acs.org/doi/abs/10.1021/bi992013z.
Hickerson, R. P.; Chepanoske, C. L.; Williams, S. D.; David, S. S.; Burrows, C. J., Mechanism-Based DNA-Protein Cross-Linking of MutY via Oxidation of 8-Oxoguanosine. J. Am. Chem. Soc. 1999, 121 (42), 9901-9902. http://pubs.acs.org/doi/abs/10.1021/ja9923484.
Golinelli, M.-P.; Chmiel, N. H.; David, S. S., Site-Directed Mutagenesis of the Cysteine Ligands to the [4Fe-4S] Cluster of Escherichia coli MutY. Biochemistry. 1999, 38 (22), 6997-7007. http://pubs.acs.org/doi/abs/10.1021/bi982300n.
Chepanoske, C. L.; Porello, S. L.; Fujiwara, T.; Sugiyama, H.; David, S. S., Substrate recognition by Escherichia coli MutY using substrate analogs. Nucleic Acids Res. 1999, 27 (15), 3197-3204. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC148548/.
Williams, S. D.; David, S. S., Evidence that MutY is a monofunctional glycosylase capable of forming a covalent Schiff base intermediate with substrate DNA. Nucleic Acids Res. 1998, 26 (22), 5123-5133. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC147954/.
Porello, S. L.; Leyes, A. E.; David, S. S., Single-Turnover and Pre-Steady-State Kinetics of the Reaction of the Adenine Glycosylase MutY with Mismatch-Containing DNA Substrates. Biochemistry. 1998, 37 (42), 14756-14764. http://pubs.acs.org/doi/abs/10.1021/bi981594%2B.
Porello, S. L.; Cannon, M. J.; David, S. S., A Substrate Recognition Role for the [4Fe-4S]2+ Cluster of the DNA Repair Glycosylase MutY. Biochemistry. 1998, 37 (18), 6465-6475. http://pubs.acs.org/doi/abs/10.1021/bi972433t.
David, S. S.; Williams, S. D., Chemistry of Glycosylases and Endonucleases Involved in Base-Excision Repair. Chem Rev. 1998, 98 (3), 1221-1262. http://pubs.acs.org/doi/abs/10.1021/cr980321h?journalCode=chreay.
Porello, S. L.; Williams, S. D.; Chepanoske, C. L.; David, S. S., Mismatch repair by the [4Fe-4S] cluster containing DNA repair enzyme, MutY. J. Inorg. Biochem. 1997, 67 (1-4), 256. https://doi.org/10.1016/S0162-0134(97)80131-6.
Porello, S. L.; Williams, S. D.; Kuhn, H.; Michaels, M. L.; David, S. S., Specific Recognition of Substrate Analogs by the DNA Mismatch Repair Enzyme MutY. J. Am. Chem. Soc. 1996, 118 (44), 10684-10692. http://pubs.acs.org/doi/abs/10.1021/ja9602206.
Eason, R. G.; Burkhardt, D. M.; Phillips, S. J.; Smith, D. P.; David, S. S., Synthesis and characterization of 8-methoxy-2′- deoxyadenosine-containing oligonucleotides to probe the syn glycosidic conformation of 2′-deoxyadenosine within DNA. Nucleic Acids Res. 1996, 24 (5), 890-7. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC145706/.
Kuhn, H.; Smith, D. P.; David, S. S., Efficient Synthesis of 2′-Deoxyformycin A Containing Oligonucleotides and Characterization of Their Stability in Duplex DNA. J. Org. Chem. 1995, 60 (22), 7094-5. http://pubs.acs.org/doi/abs/10.1021/jo00127a010.