
Kori Lay
Education:
B.A. Chemistry and Environmental Studies, UC Santa Barbara 2014
From: Port Washington, New York
Joined the David Lab: January 2016
Outside of lab: I love doing outdoor activities like hiking, camping,
snowboarding. I also enjoy cooking and make a mean red Thai curry.
Research in David Lab:
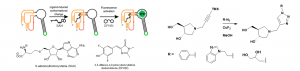
My work in the David lab focuses on studying methylthioadenosine nucleosidase (MTAN), which is an enzyme found in bacteria. This enzyme is of interest to us because it is structurally similar to the human enzyme, methylthioadenosine phosphorylase (MTAP), which is an anticancer target. MTAN is also a potential antibiotic target due to it’s involvement in quorum sensing in bacteria. S-adenosylhomocysteine (SAH) is a substrate of MTAN and the transition state formed between MTAN and SAH has been used in the development of tight binding inhibitors of MTAN. Potent inhibitors of MTAN have been developed but their synthesis is arduous and long. Using Copper-Catalyzed Azide-Alkyne Cycloaddition (CuAAC), I have synthesized potential inhibitors of MTAN quickly and efficiently (shown below). Using an RNA-based fluorescent biosensor developed in the Hammond lab, I have designed an in vitro assay for detecting MTAN activity and inhibition that is easier and more sensitive than previous activity assays.1 Using this assay, I will be testing for inhibition of MTAN using the CuAAC synthesized inhibitors. This assay also works in vivo, so inhibitors will also be tested in E. coli.
Links to Papers from David Lab:
Previous Research Experience:
As an undergraduate, I did research in Dr. Daniel Morse’s lab where I worked to produce transmission electron microscopy images of the cross section of iridocyte cells found in Tridacnid giant clams.2 My aim was to analyze the structure-function relationships of the internal Bragg-reflectors in these cells, to better understand the mechanism of their newly discovered enhancement of photosynthesis by the endosymbiotic algae living within the clam’s tissues.
- Su, Y.; Hickey, S. F.; Keyser, S. G. L.; Hammond, M. C., In Vitro and In Vivo Enzyme Activity Screening via RNA-Based Fluorescent Biosensors for S-Adenosyl-l-homocysteine (SAH). Journal of the American Chemical Society 2016, 138 (22), 7040-7047.
- Ghoshal, A.; Eck, E.; Morse, D. E., Biological analogs of RGB pixelation yield white coloration in giant clams. Optica 2016, 3 (1), 108-111.
