New Manuscript Published: Selective base excision repair of DNA damage by the non‐base‐flipping DNA glycosylase AlkC.
The preservation of genomic integrity performed by DNA repair machinery is crucial for living organisms, and malfunctions in DNA repair machinery can have far-reaching and devastating effects on a cell’s ability to attain precise DNA replication, properly regulate cell differentiation and self-renewal, and to regulate cell growth and apoptosis, among other important cellular functions. Mutations of critical residues in DNA repair proteins can drastically reduce DNA repair capability in cells, allowing for a build-up of genomic mutations. Inherited variants in DNA repair proteins such as glycosylase MUTYH have been linked to a predisposition to tumors in patients with disease MUTYH Associated Polyposis (MAP). The David Lab is interested in delineating DNA repair mechanisms to help shed light on the etiology of cancer and other diseases, providing mechanistic and structural information that may be used, for example, to design drug molecules targeting DNA repair proteins.
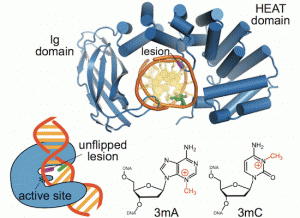
New work from the David Lab examined the selective base excision repair of DNA damage by the non-base-flipping DNA glycosylase AlkC, which primarily targets alkylated-DNA damage product N3-methyladenine (3mA). This work details how AlkC selects for and excises 3mA with its non-base-flipping mechanism. The authors carried out a comprehensive phylogenetic, biochemical, and structural comparison of AlkC and AlkD proteins for comparison, which shows, notably, characteristics important for substrate specificity and why bulkier substrates are not preferred. Interestingly, AlkC’s excision mechanism involves using HEAT-like repeat domains and in most cases Ig-like domains to introduce a kink in the target DNA, helping to expose the target nucleobase, allowing for subsequent insertion of the enzyme active site to excise its target.
Click here to read more about AlkC’s non-base-flipping mechanism.
Source:
DOI 10.15252/embj.201797833| Published online 20.10.2017
The EMBO Journal (2017) e201797833













