Recent Article Published by Sheila David’s Lab: Unique Hydrogen Bonding of Adenine with the Oxidatively Damaged Base 8-Oxoguanine Enables Specific Recognition and Repair by DNA Glycosylase MutY.
Majumdar, C.; Mckibbin, P.L.; Krajewski, A.E.; Manlove, A.H.; Lee, J.K.; David, S.S.
J. Am. Soc. 2020. 142, 48, 20340–20350.
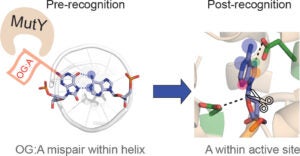
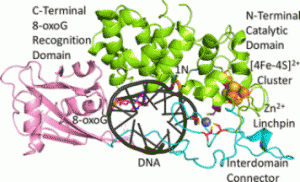
DNA repair protein MutY employs specific interactions to differentiate OG:A basepairs from canonical G:C and T:A basepairs. Prior work from our lab has focused on understanding the structural requirements of OG on lesion recognition and catalysis, and we have shown that MutY relies on the exocyclic 2-amino group of OG to identify and distinguish OG:A from other basepairs. Additionally, we’ve shown that OG binding induces conformational changes that influence A excision.
This new work uses structure-activity relationships (SARs) to identify the structural features of A that influence OG:A recognition, verification, base excision, and overall cellular repair. We correlate observed in vitro MutY activity on A analogue substrates with their experimental and calculated acidities to provide mechanistic insight into the factors influencing MutY base excision efficiency. Our results herein can be used to guide future design of MutY/MUTYH specific probes to monitor the activity, or lack thereof, of MutY/MUTYH variants. These results can also applied toward the development of MUTY/MUTYH specific inhibitors that may find utility in cancer therapeutics.
Click on the link or graphical abstract to find out more!
https://pubs.acs.org/doi/abs/10.1021/jacs.0c06767#